Marvin Martens, Ammar Ammar, Anders Riutta, Andra Waagmeester, Denise N Slenter, Kristina Hanspers, Ryan A. Miller, Daniela Digles, Elisson N Lopes, Friederike Ehrhart, Lauren J Dupuis, Laurent A Winckers, Susan L Coort, Egon L Willighagen, Chris T Evelo, Alexander R Pico, Martina Kutmon, WikiPathways: connecting communities, Nucleic Acids Research, , gkaa1024, https://doi.org/10.1093/nar/gkaa1024
Abstract
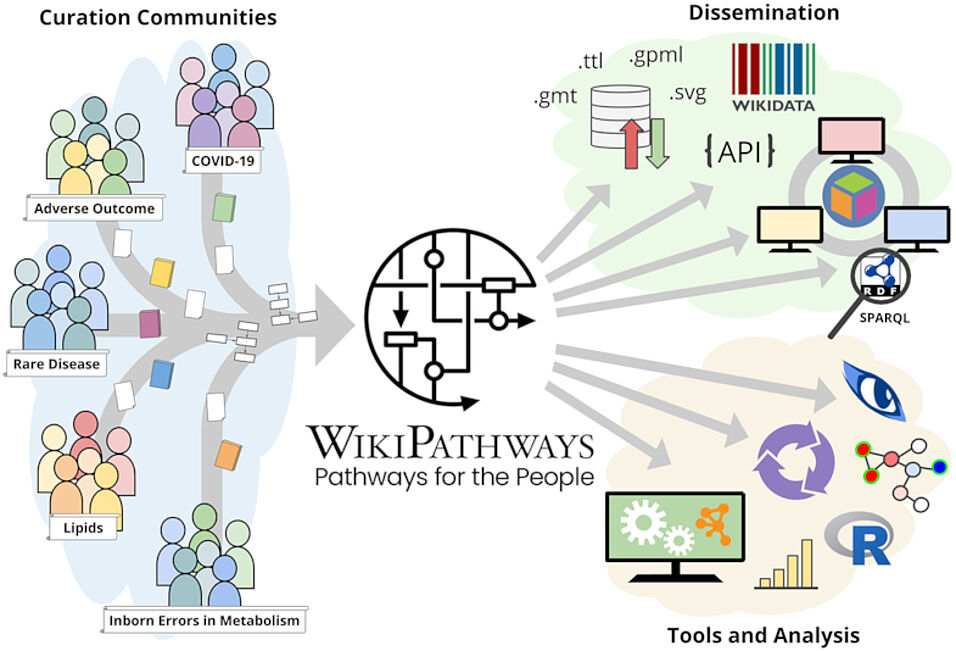
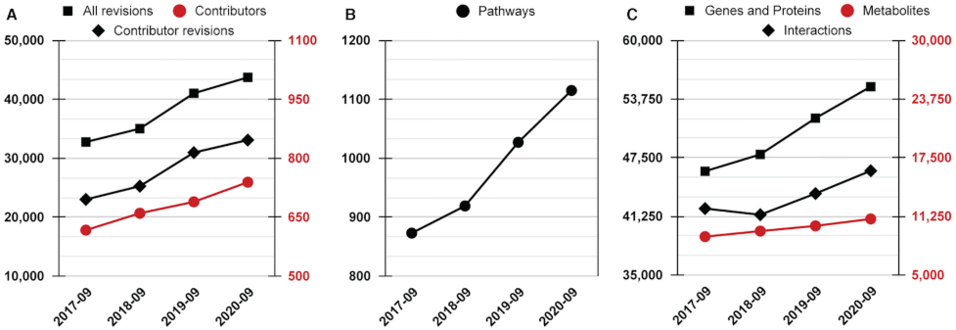
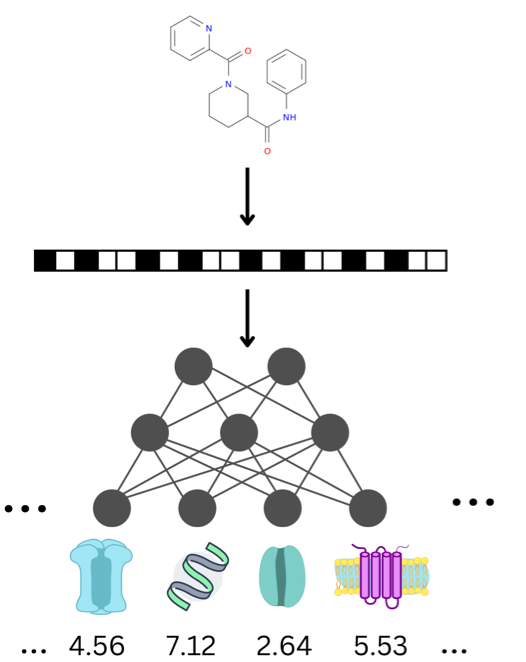
WikiPathways (https://www.wikipathways.org) is a biological pathway database known for its collaborative nature and open science approaches. With the core idea of the scientific community developing and curating biological knowledge in pathway models, WikiPathways lowers all barriers for accessing and using its content. Increasingly more content creators, initiatives, projects and tools have started using WikiPathways. Central in this growth and increased use of WikiPathways are the various communities that focus on particular subsets of molecular pathways such as for rare diseases and lipid metabolism. Knowledge from published pathway figures helps prioritize pathway development, using optical character and named entity recognition. We show the growth of WikiPathways over the last three years, highlight the new communities and collaborations of pathway authors and curators, and describe various technologies to connect to external resources and initiatives. The road toward a sustainable, community-driven pathway database goes through integration with other resources such as Wikidata and allowing more use, curation and redistribution of WikiPathways content.
Acknowledgements and funding
We would like to thank everyone who contributed to WikiPathways content, code and discussions, and who used the content of WikiPathways in their projects.
Author contributions: M.M. and M.K. led the writing of the manuscript. All authors contributed to the writing of the manuscript. Middle authors are sorted alphabetically by the first name and based on their role within the project (core developers, curation team), followed by principal investigators.
European Union’s Horizon 2020 research and innovation programme [NanoCommons: 731032 to L.W., E.L.W., NanoSolveIT: 814572 to A.A., E.L.W., RiskGONE: 814425 to A.A., E.L.W., OpenRiskNet: 731075 to M.M., C.T.E., EJP-RD: 825575 to F.E., C.T.E.]; ZonMw [10430012010015 to M.K., C.T.E, S.L.C, F.E.]; National Institute of General Medical Sciences [R01 GM100039 to A.R., K.H., A.R.P., R01 GM089820 to A.W.].
Rights and permissions
This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted reuse, distribution, and reproduction in any medium, provided the original work is properly cited.