Brecklinghaus T, Albrecht W, Kappenberg F, Duda J, Vartak N, Edlund K, Marchan R, Ghallab A, Cadenas C, Günther G, Leist M, Zhang M, Gardner I, Reinders J, Russel FG, Foster AJ, Williams DP, Damle-Vartak A, Grandits M, Ecker G, Kittana N, Rahnenführer J, Hengstler JG. The hepatocyte export carrier inhibition assay improves the separation of hepatotoxic from non-hepatotoxic compounds. Chem Biol Interact. 2021 Oct 27:109728.
DOI
https://doi.org/10.1016/j.cbi.2021.109728
Abstract
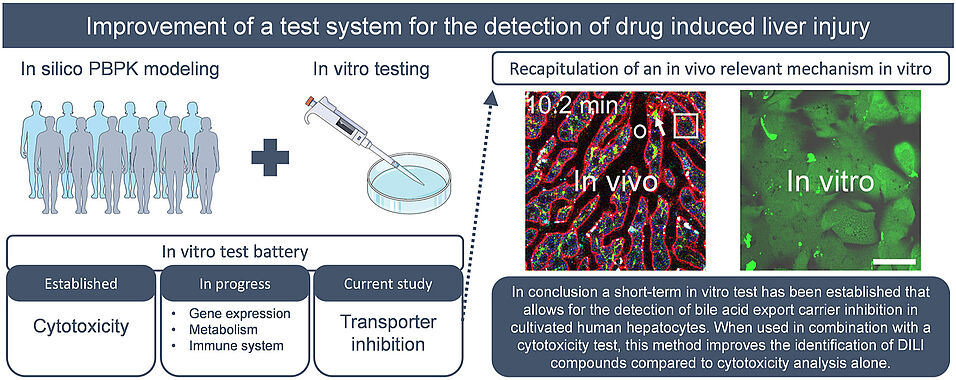
An in vitro/in silico method that determines the risk of human drug induced liver injury in relation to oral doses and blood concentrations of drugs was recently introduced. This method utilizes information on the maximal blood concentration (Cmax) for a specific dose of a test compound, which can be estimated using physiologically-based pharmacokinetic modelling, and a cytotoxicity test in cultured human hepatocytes. In the present study, we analyzed if the addition of an assay that measures the inhibition of bile acid export carriers, like BSEP and/or MRP2, to the existing method improves the differentiation of hepatotoxic and non-hepatotoxic compounds. Therefore, an export assay for 5-chloromethylfluorescein diacetate (CMFDA) was established. We tested 36 compounds in a concentration-dependent manner for which the risk of hepatotoxicity for specific oral doses and the capacity to inhibit hepatocyte export carriers are known. Compared to the CTB cytotoxicity test, substantially lower EC10 values were obtained using the CMFDA assay for several known BSEP and/or MRP2 inhibitors. To quantify if the addition of the CMFDA assay to our test system improves the overall separation of hepatotoxic from non-hepatotoxic compounds, the toxicity separation index (TSI) was calculated. We obtained a better TSI using the lower alert concentration from either the CMFDA or the CTB test (TSI: 0.886) compared to considering the CTB test alone (TSI: 0.775). In conclusion, the data show that integration of the CMFDA assay with an in vitro test battery improves the differentiation of hepatotoxic and non-hepatotoxic compounds in a set of compounds that includes bile acid export carrier inhibitors.
Funding
This work has been supported in part by the Research Training Group “Biostatistical Methods for High-Dimensional Data in Toxicology” (RTG 2624, Project P1 and P2) funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation - Project Number 427806116). Furthermore, this work has received funding from the European Union's Horizon 2020 research and innovation programme under grant agreement no. 681002 (EU-ToxRisk) and LivSysTransfer (BMBF, 031L0119).
Copyright
© 2021. Brecklinghaus T, Albrecht W, Kappenberg F, Duda J, Vartak N, Edlund K, Marchan R, Ghallab A, Cadenas C, Günther G, Leist M, Zhang M, Gardner I, Reinders J, Russel FG, Foster AJ, Williams DP, Damle-Vartak A, Grandits M, Ecker G, Kittana N, Rahnenführer J, Hengstler JG. The hepatocyte export carrier inhibition assay improves the separation of hepatotoxic from non-hepatotoxic compounds. Chem Biol Interact. 2021 Oct 27:109728. This is an open access article distributed under the terms of the Creative Commons CC-BY license, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords
Cholestasis, DILI, Transport