Sosnin, S. MolCompass: multi-tool for the navigation in chemical space and visual validation of QSAR/QSPR models. J Cheminform 16, 98 (2024).
DOI
https://doi.org/10.1186/s13321-024-00888-z
Abstract
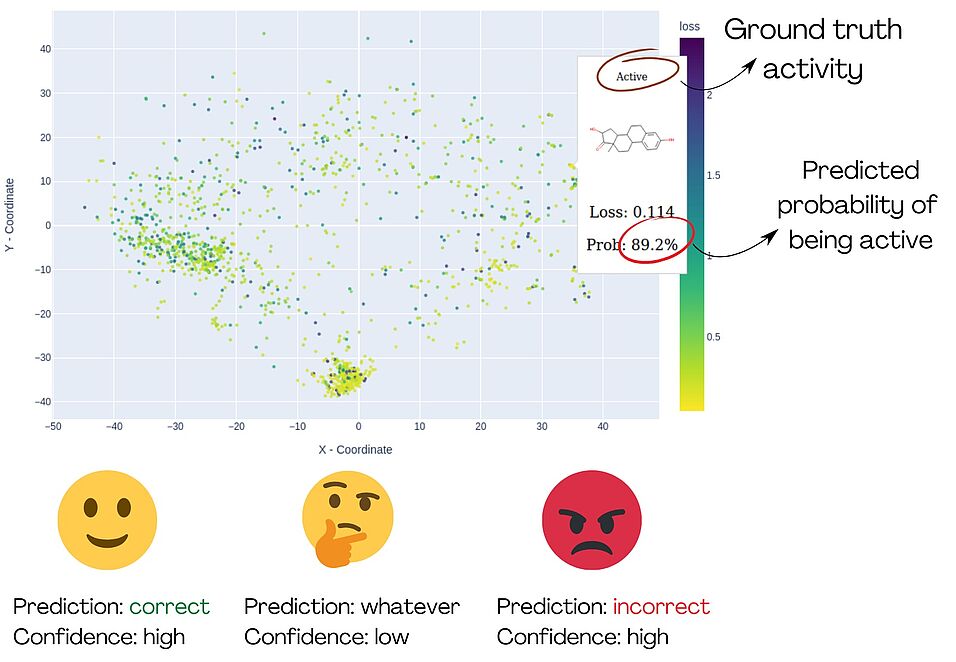
The exponential growth of data is challenging for humans because their ability to analyze data is limited. Especially in chemistry, there is a demand for tools that can visualize molecular datasets in a convenient graphical way. We propose a new, ready-to-use, multi-tool, and open-source framework for visualizing and navigating chemical space. This framework adheres to the low-code/no-code (LCNC) paradigm, providing a KNIME node, a web-based tool, and a Python package, making it accessible to a broad cheminformatics community. The core technique of the MolCompass framework employs a pre-trained parametric t-SNE model. We demonstrate how this framework can be adapted for the visualisation of chemical space and visual validation of binary classification QSAR/QSPR models, revealing their weaknesses and identifying model cliffs. All parts of the framework are publicly available on GitHub, providing accessibility to the broad scientific community.
Scientific contribution
We provide an open-source, ready-to-use set of tools for the visualization of chemical space. These tools can be insightful for chemists to analyze compound datasets and for the visual validation of QSAR/QSPR models.
Press Release
Read here the official press release of the University of Vienna: 'Molecular Compass' points way to Reduction of Animal Testing - Scientists Develop Smart Software Tool for Chemical Risk Evaluation
Funding
This work was supported by the project RISK-HUNT3R: RISK assessment of chemicals integrating HUman centric Next generation Testing strategies promoting the 3Rs. RISK-HUNT3R has received funding from the European Union's Horizon 2020 research and innovation programme under grant agreement No 964537 and is part of the ASPIS cluster.
Rights & permissions
This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.