Huang J, Osthushenrich T, MacNamara A, Mälarstig A, Brocchetti S, Bradberry S, Scarabottolo L, Ferrada E, Sosnin S, Digles D, Superti-Furga G, Ecker GF (2024). ProteoMuaMetrics: Machine Learning Approaches for Solute Carrier Family 6 Mutation Pathogenicity Prediction. RSC Advances 14:13083-13094
DOI
https://doi.org/10.1039/d4ra00748d
Abstract
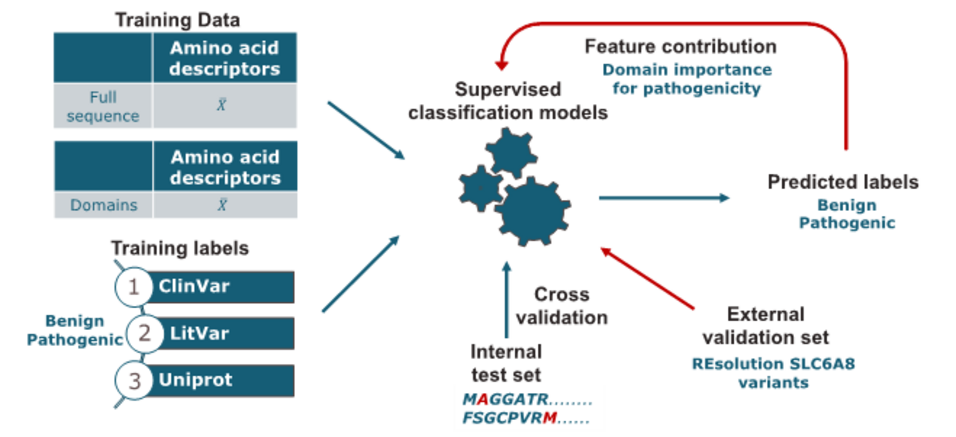
The solute carrier transporter family 6 (SLC6) is of key interest for their critical role in the transport of small amino acids or amino acid-like molecules. Their dysfunction is strongly associated with human diseases such as including schizophrenia, depression, and Parkinson's disease. Linking single point mutations to disease may support insights into the structure–function relationship of these transporters. This work aimed to develop a computational model for predicting the potential pathogenic effect of single point mutations in the SLC6 family. Missense mutation data was retrieved from UniProt, LitVar, and ClinVar, covering multiple protein-coding transcripts. As encoding approach, amino acid descriptors were used to calculate the average sequence properties for both original and mutated sequences. In addition to the full-sequence calculation, the sequences were cut into twelve domains. The domains are defined according to the transmembrane domains of the SLC6 transporters to analyse the regions' contributions to the pathogenicity prediction. Subsequently, several classification models, namely Support Vector Machine (SVM), Logistic Regression (LR), Random Forest (RF), and Extreme Gradient Boosting (XGBoost) with the hyperparameters optimized through grid search were built. For estimation of model performance, repeated stratified k-fold cross-validation was used. The accuracy values of the generated models are in the range of 0.72 to 0.80. Analysis of feature importance indicates that mutations in distinct regions of SLC6 transporters are associated with an increased risk for pathogenicity. When applying the model on an independent validation set, the performance in accuracy dropped to averagely 0.6 with high precision but low sensitivity scores.
Funding
This work was performed within the REsolution project. REsolution has received funding from the Innovative Medicines Initiative 2 Joint Undertaking (https://ihi.europa.eu) under grant agreement no. 101034439. This Joint Undertaking receives support from the European Union's Horizon 2020 Research and Innovation Programme and EFPIA. This article reflects only the authors' views and neither IMI nor the European Union and EFPIA are responsible for any use that may be made of the information contained therein. This work was also supported by the Austrian Science Fund/FWF, grant W1232 (MolTag).
Rights & permissions
This is an open access article distributed under the terms of the Creative Commons CC-BY license, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.