Smajić, A., Grandits, M. & Ecker, G.F. Using Jupyter Notebooks for re-training machine learning models. J Cheminform 14, 54 (2022).
DOI
https://doi.org/10.1186/s13321-022-00635-2
Abstract
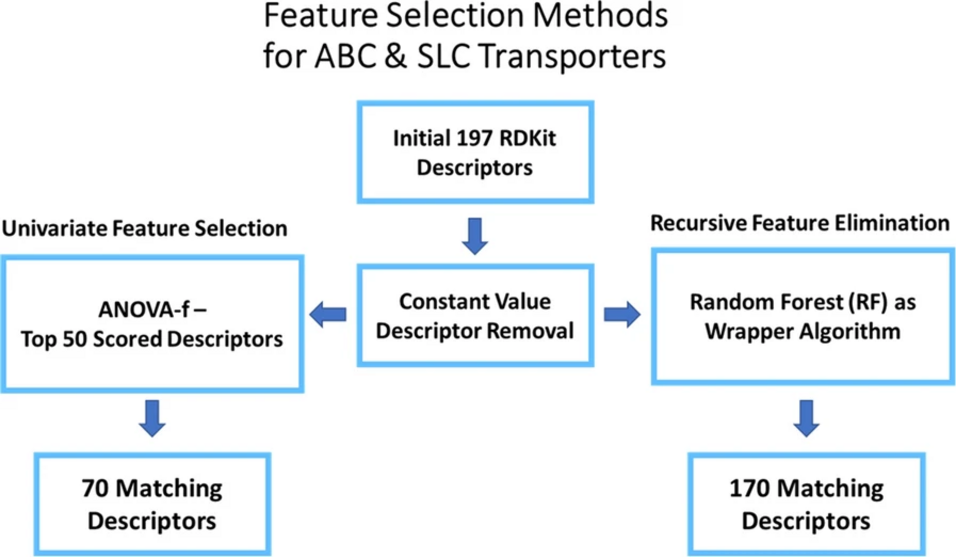
Machine learning (ML) models require an extensive, user-driven selection of molecular descriptors in order to learn from chemical structures to predict actives and inactives with a high reliability. In addition, privacy concerns often restrict the access to sufficient data, leading to models with a narrow chemical space. Therefore, we propose a framework of re-trainable models that can be transferred from one local instance to another, and further allow a less extensive descriptor selection. The models are shared via a Jupyter Notebook, allowing the evaluation and implementation of a broader chemical space by keeping most of the tunable parameters pre-defined. This enables the models to be updated in a decentralized, facile, and fast manner. Herein, the method was evaluated with six transporter datasets (BCRP, BSEP, OATP1B1, OATP1B3, MRP3, P-gp), which revealed the general applicability of this approach.
Funding
This project has received funding from the Innovative Medicines Initiative 2 Joint Undertaking under grant agreement No 777365 (eTRANSAFE). This Joint Undertaking receives support from the European Union’s Horizon 2020 research and innovation programme and EFPIA. The Pharmacoinformatics Research Group (Ecker lab) acknowledges funding provided by the Austrian Science Fund FWF AW012321 MolTag.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.