S. Decker, A. Taschauer, E. Geppl, V. Pirhofer, M. Schauer, S. Pöschl, F. Kopp, L. Richter, G.F. Ecker, H. Sami, M. Ogris, Structure-based peptide ligand design for improved epidermal growth factor receptor targeted gene delivery, European Journal of Pharmaceutics and Biopharmaceutics (2022)
DOI
https://doi.org/10.1016/j.ejpb.2022.05.004
Abstract
The epidermal growth factor receptor EGFR allows targeted delivery of macromolecular drugs to tumors. Its ligand, epidermal growth factor, binds EGFR with high affinity but acts mitogenic. Non-mitogenic peptides are utilized as targeting ligands, like the dodecapeptide GE11, although its low binding affinity warrants improvement.
We applied a two-step computational approach with database search and molecular docking to design GE11 variants with improved binding. Synthesized peptides underwent binding studies on immobilized EGFR using surface plasmon resonance. Conjugates of peptides coupled via heterobifunctional PEG linker to linear polyethylenimine (LPEI) were used for transfection studies on EGFR-overexpressing cells using reporter gene encoding plasmid DNA.
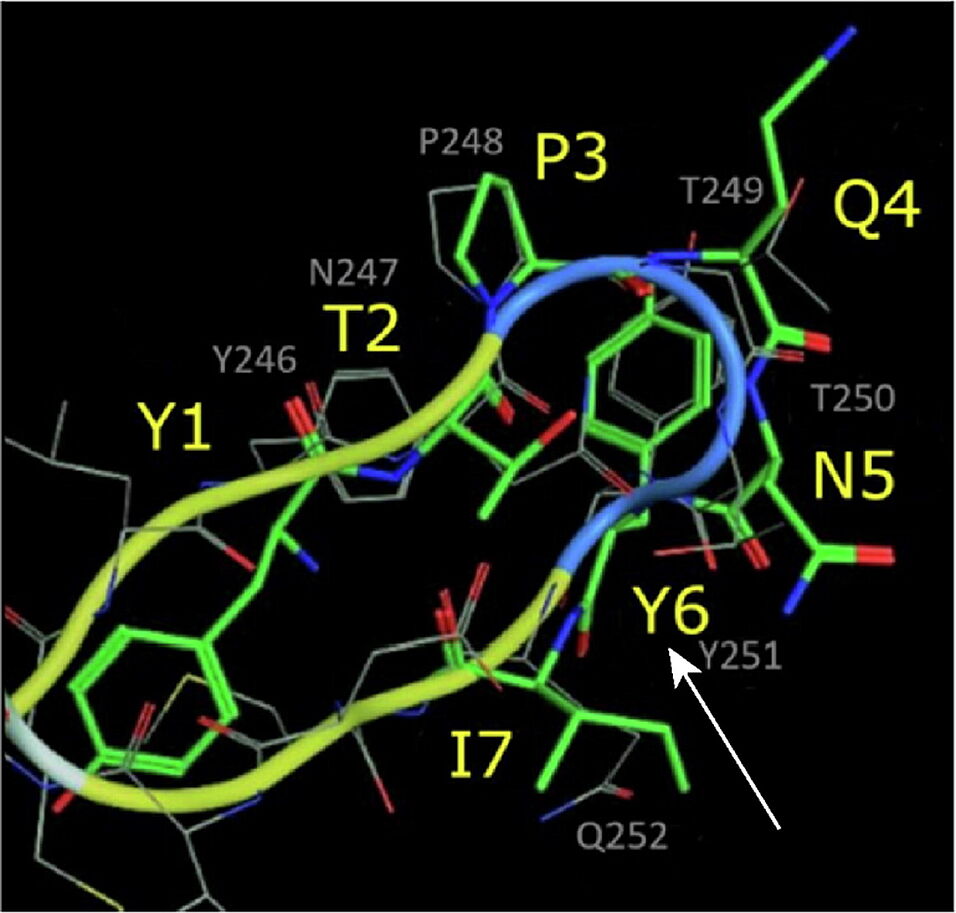
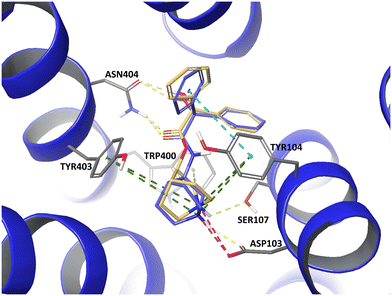
Docking studies unraveled similarities between GE11 and the EGFR dimerization arm. By skipping non-overlapping amino acids, a less hydrophobic segment (YTPQNVI) was identified being directly involved in EGFR binding. By replacing valine by tyrosine, a full-length version with proposed enhanced binding (GE11m3) was developed. While hydrophobic or hydrophilic segments and variations thereof exhibited low binding, GE11m3 exhibited 3-fold increase in binding compared to GE11, validating in silico predictions. In transfection studies, polyplexes with GE11m3 induced a significantly higher reporter gene expression when compared to GE11 polyplexes both on murine and human cancer cells overexpressing EGFR.
Funding
This work was in part co-funded by a grant from the Austrian Research Promotion Agency FFG. Open access funding was provided by the University of Vienna.
Rights and Permissions
This is an open access article distributed under the terms of the Creative Commons CC-BY license, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords
Polyplex, transfection, targeted gene delivery, EGFR, GE11