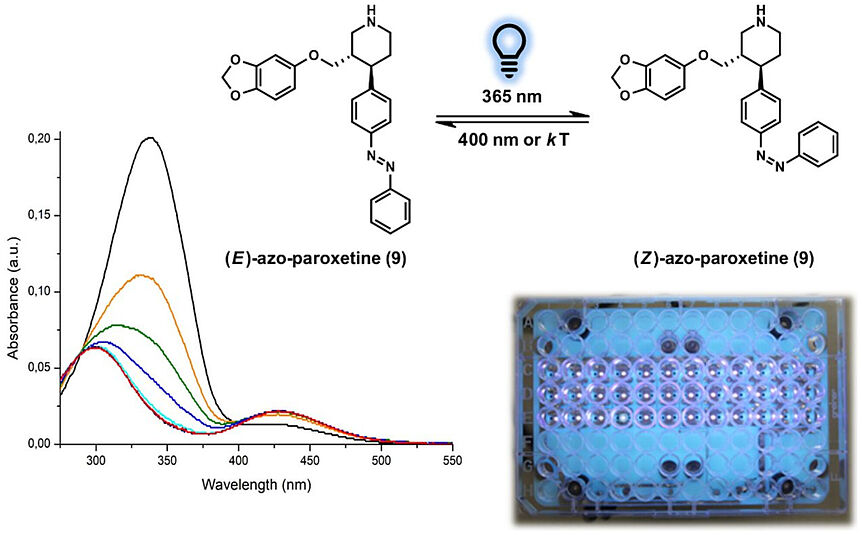
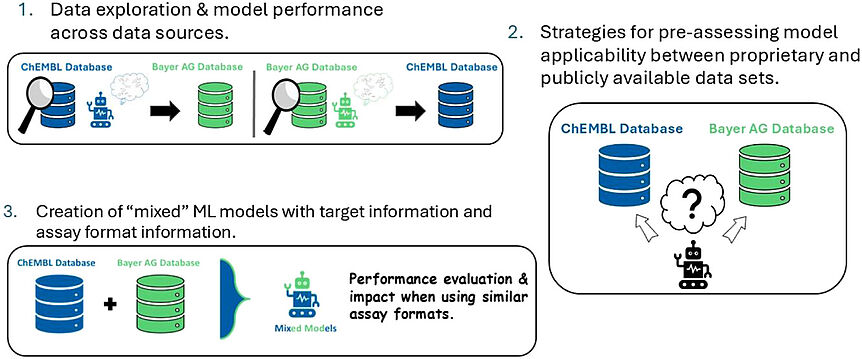
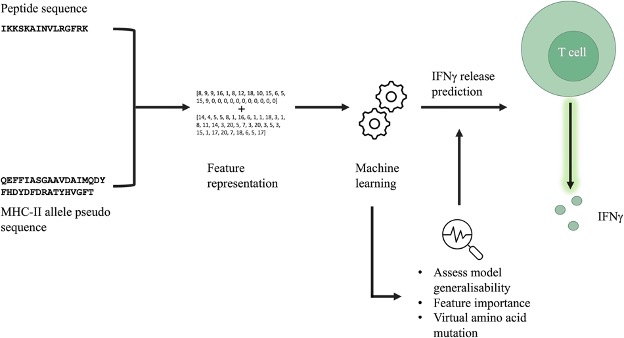
The 4th consortium meeting was planned and scheduled in Paris - but then COVID-19 happened. As a result the RESOLUTE project meeting in person was cancelled of course...
BUT although the RESOULTE consortium couldn't meet in person this time, they instead tried to make this virtual meeting special and so they asked every participant for a French-themed picture contribution for the virtual "group picture". The result you can see below or on your right hand side.
The speakers also made an extra effort to tailor their talks to the virtual visit in Paris. The first session for example was held just next to the famous Eiffel Tower and later continued at the Arc de Triomphe. Just amazing!
As a second anniversary present we are also happy to share the RESOLUTE commentary officially published in Nature Reviews Drug Discovery just beginning of July. Congratulations!
The RESOLUTE consortium: unlocking SLC transporters for drug discovery
We again want to send a big "thank you" to the organisers for this very nice virtual visit to Paris! Well done.