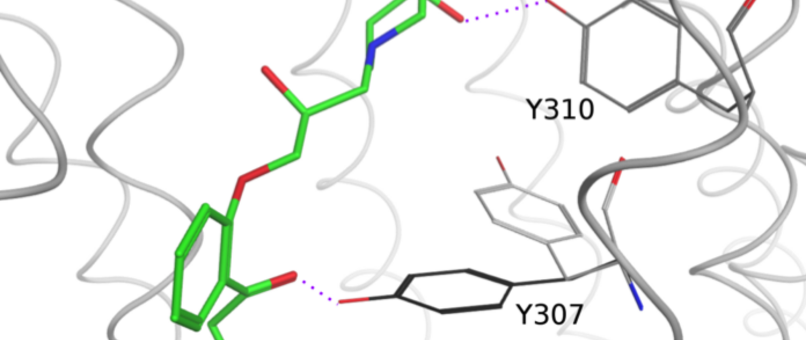
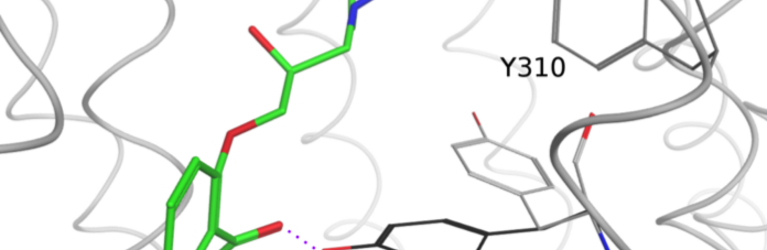
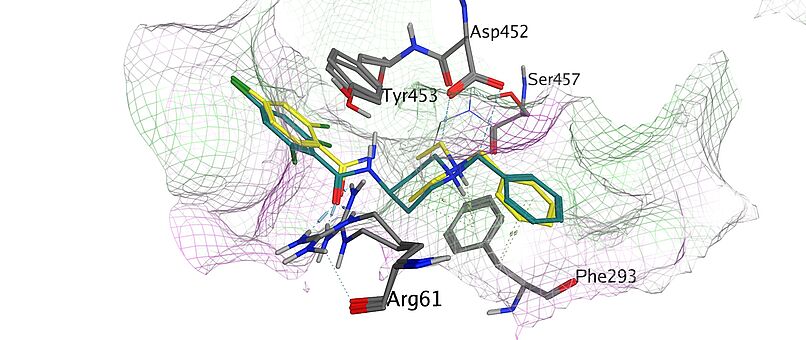
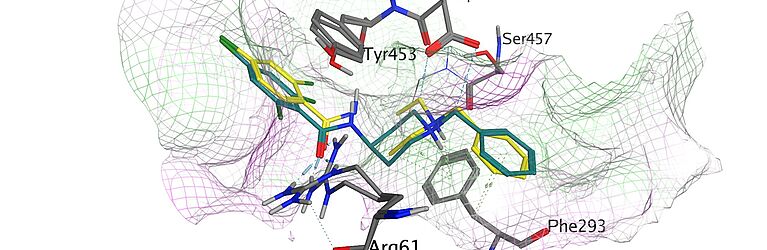
Mission Statement
Following a holistic pharmacoinformatic approach we combine structural modeling of proteins, structure-based drug design, chemometric and in silico chemogenomic methods, statistical modeling and machine learning approaches to develop predictive computational tools for drug discovery and development. Hereby, the main focus of our work comprise interaction of ligands with transmembrane transport proteins and prediction of toxicity. The validation and optimisation of the obtained in silico models by strong links to experimental groups is an integral part of these activities.
Furthermore, we aim at developing tools for in vitro to in vivo translation in the area of adverse drug event prediction, thereby leveraging integrated life science data. Research activities are complemented by strong educational activities, outlined in the doctoral program MolTag and in EUROPIN, a European PhD Program in Pharmacoinformatics.